5.4 PCA Proteomics Data Analysis in R/Bioconductor
Principal component analysis (PCA) in R programming is an analysis of the linear components of all existing attributes. Principal components are linear combinations (orthogonal transformation) of the original predictor in the dataset.
Principal component analysis (PCA) biplot generated in R using... Download Scientific Diagram
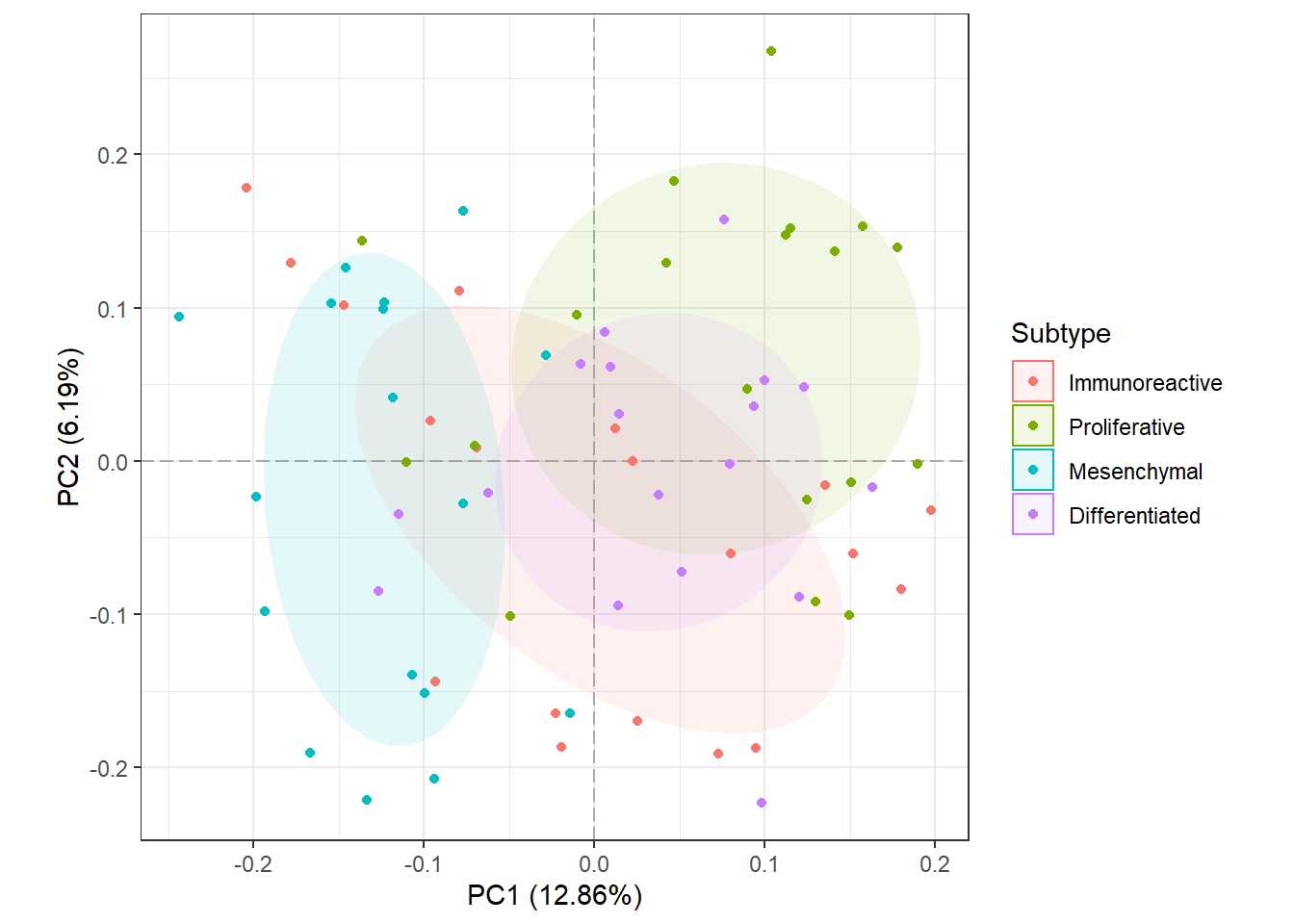
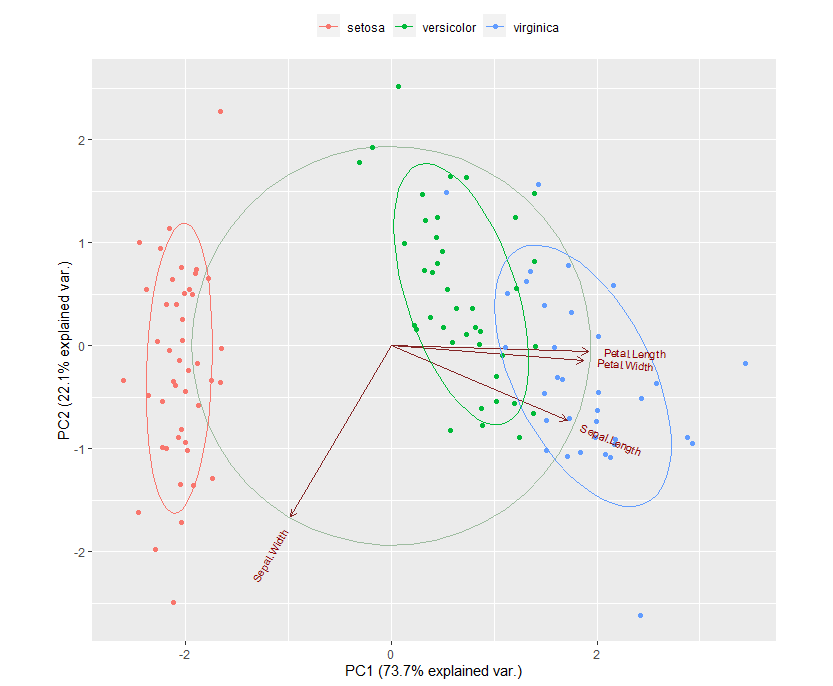
PCA is an exploratory data analysis based in dimensions reduction. The general idea is to reduce the dataset to have fewer dimensions and at the same time preserve as much information as possible. PCA allows us to make visual representations in two dimensions and check for groups or differences in the data related to different states.
Principal component analysis in R vs. R software and data mining Easy
Principal Component Analysis (PCA) is a very powerful technique that has wide applicability in data science, bioinformatics, and further afield. It was initially developed to analyse large volumes of data in order to tease out the differences/relationships between the logical entities being analysed.
Principal component analysis (PCA) in R Rbloggers
Principal components analysis, often abbreviated PCA, is an unsupervised machine learning technique that seeks to find principal components - linear combinations of the original predictors - that explain a large portion of the variation in a dataset.
Principal Component Analysis in R vs Articles STHDA
In this tutorial you'll learn how to perform a Principal Component Analysis (PCA) in R. The table of content is structured as follows: 1) Example Data & Add-On Packages 2) Step 1: Calculate Principal Components 3) Step 2: Ideal Number of Components 4) Step 3: Interpret Results 5) Video, Further Resources & Summary
Principal component analysis (PCA) in R Rbloggers
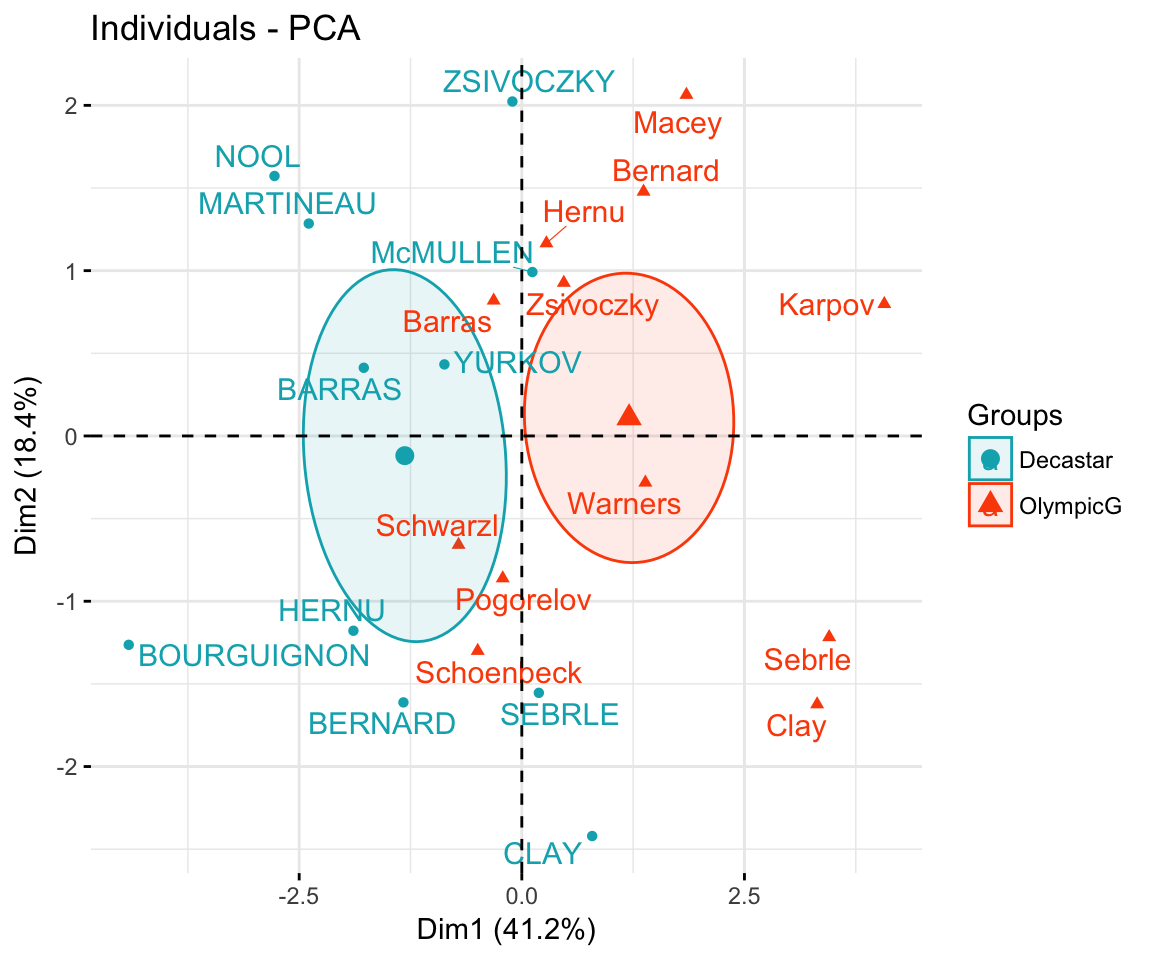
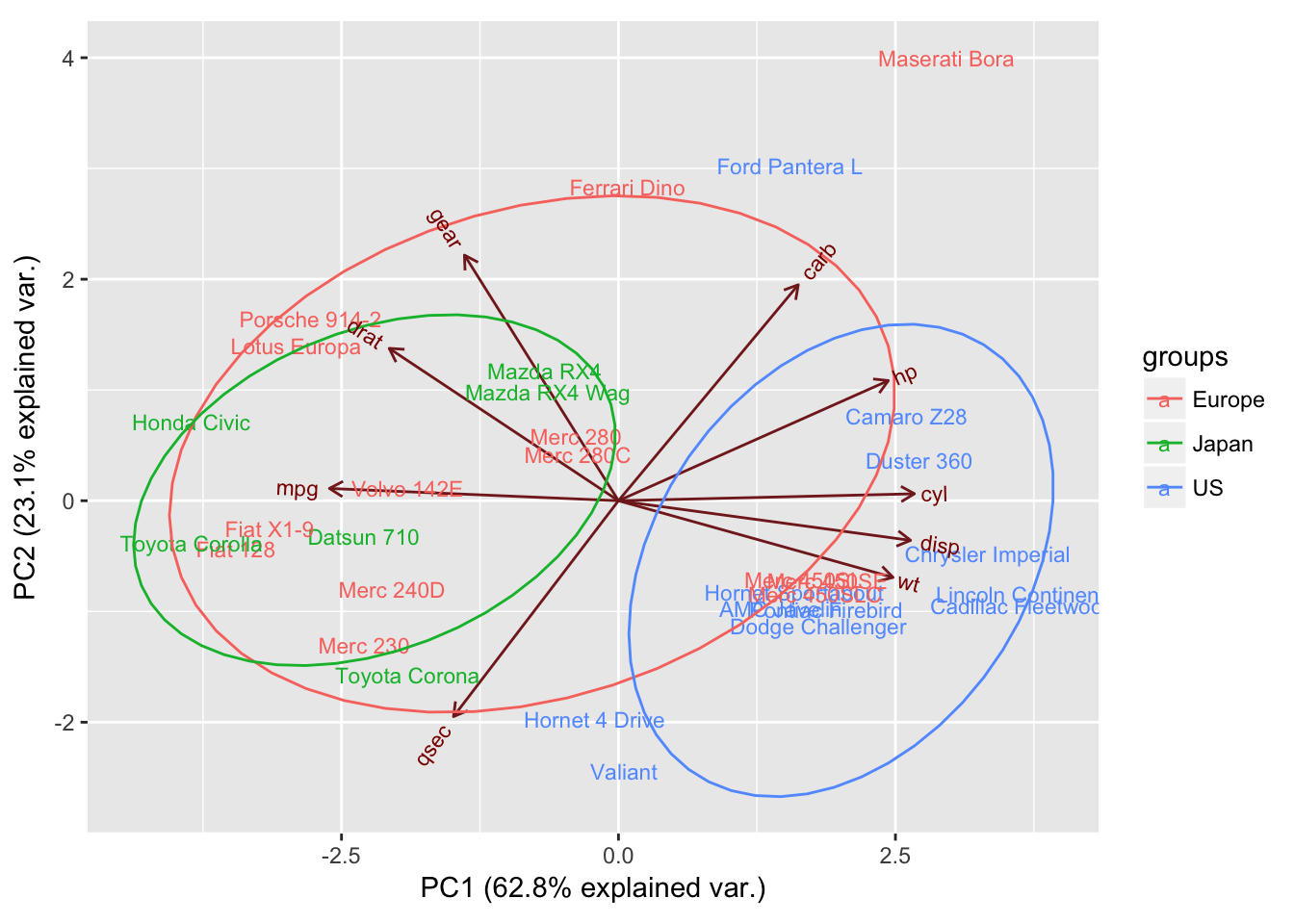
PCA Functions in R Using PC Scores The Biplot: Visualizing a PCA Conclusions References Introduction We are focusing today on Principal Components Analysis (PCA), which is an eigenanalysis-based approach. We begin, therefore, by reviewing eigenanalysis (for more details on this topic, refer to the chapter about Matrix Algebra ).

Principal component analysis in R YouTube
Contact us Principal Component Analysis (PCA) using R Posted on September 28, 2021 by Statistical Aid in R bloggers | 0 Comments [This article was first published on R tutorials - Statistical Aid: A School of Statistics, and kindly contributed to R-bloggers ].

enpca_examples [Analysis of community ecology data in R]
Principal Component Analysis (PCA) in R Tutorial | DataCamp Home About R Learn R Principal Component Analysis in R Tutorial In this tutorial, you'll learn how to use R PCA (Principal Component Analysis) to extract data with many variables and create visualizations to display that data. Updated Feb 2023 · 15 min read
R PCA Tutorial (Principal Component Analysis) DataCamp
PCA is commonly used as one step in a series of analyses. You can use PCA to reduce the number of variables and avoid multicollinearity, or when you have too many predictors relative to the number of observations. tl;dr This tutorial serves as an introduction to Principal Component Analysis (PCA). 1
Principal Component Analysis in R vs Articles STHDA
PCA is used in exploratory data analysis and for making decisions in predictive models. PCA commonly used for dimensionality reduction by using each data point onto only the first few principal components (most cases first and second dimensions) to obtain lower-dimensional data while keeping as much of the data's variation as possible.
Apply Principal Component Analysis in R (PCA Example & Results)
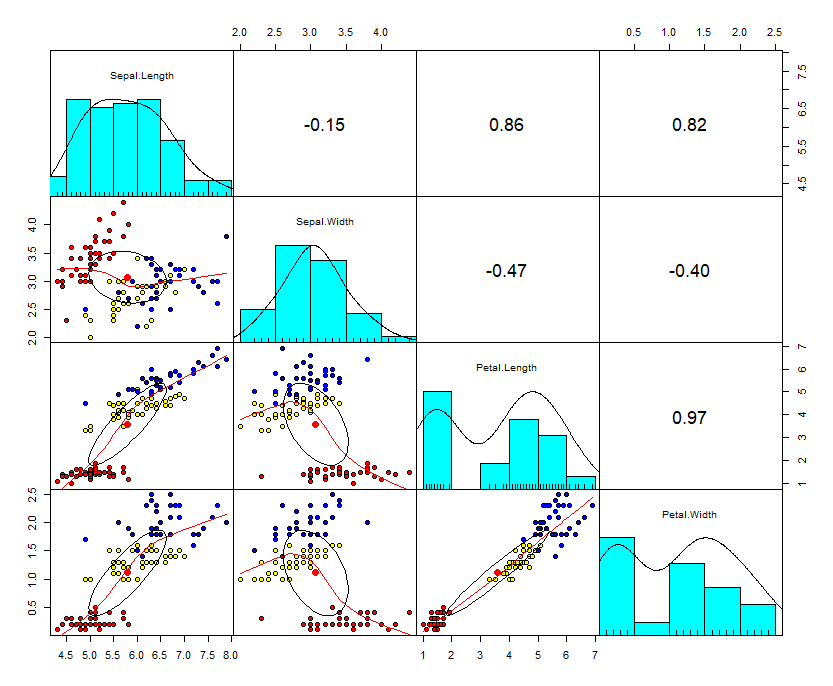
PCA of a covariance matrix can be computed as svd of unscaled, centered, matrix. Center a matrix Recall we had two vector x_obs, y_obs. We can center these columns by subtracting the column mean from each object in the column. We can perform PCA of the covariance matrix is several ways. SVD of the centered matrix.
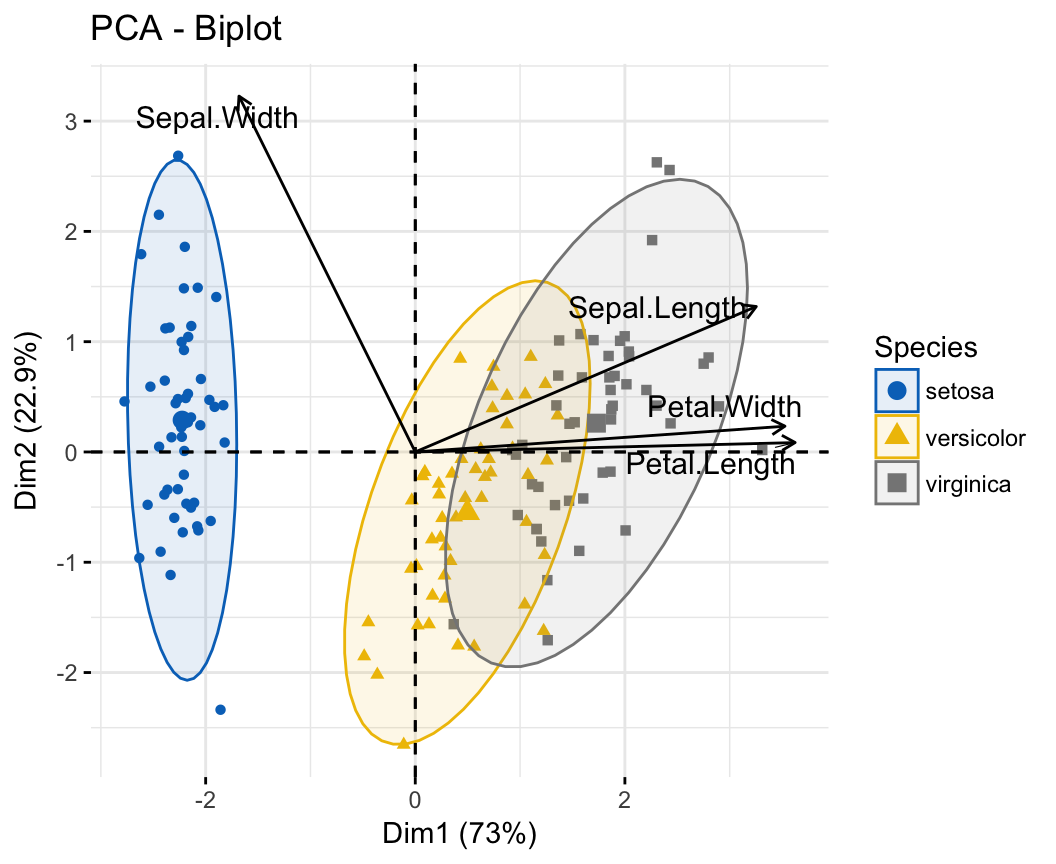
PCA Principal Component Analysis Essentials Articles STHDA
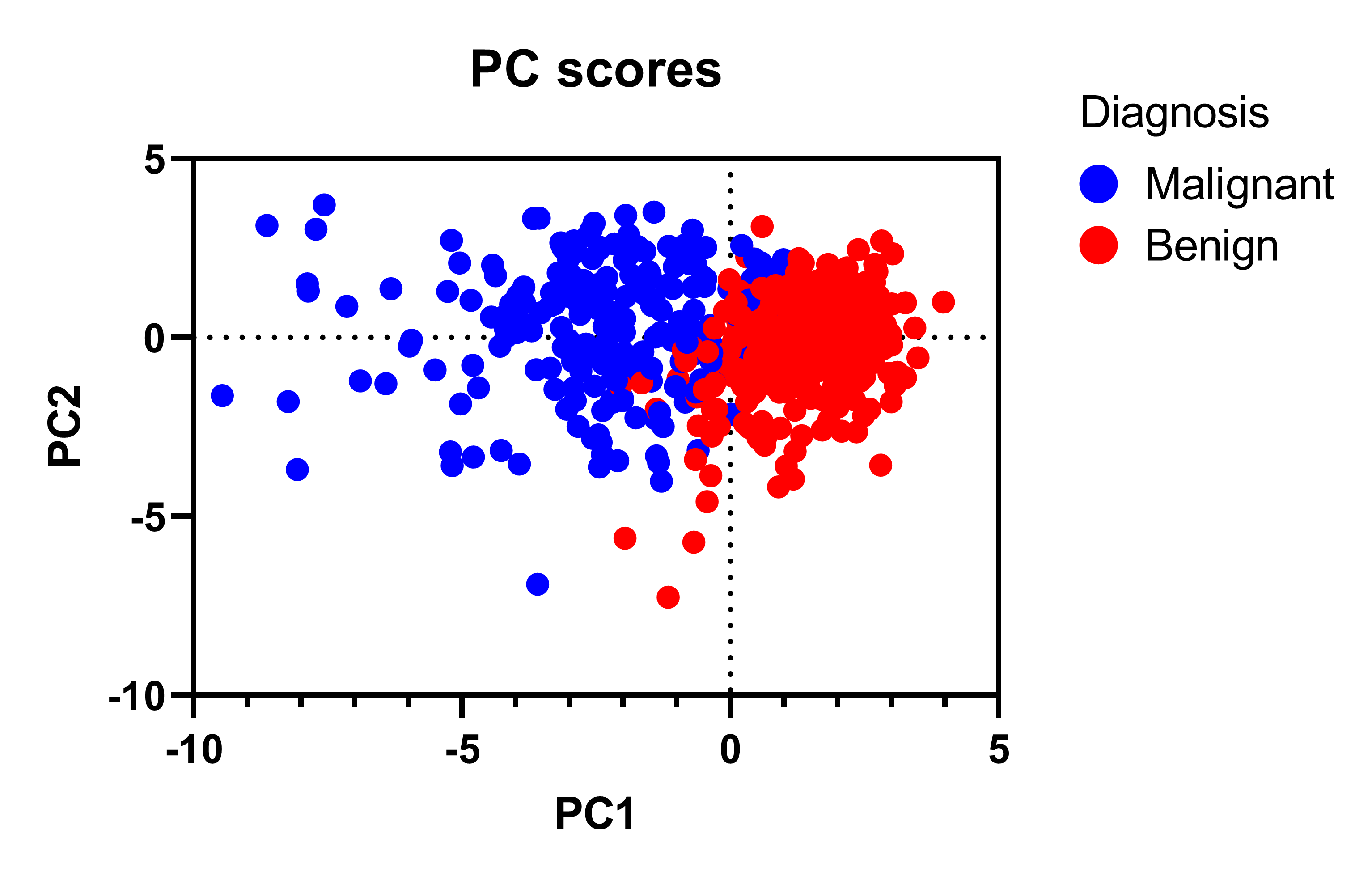
Principal Component Analysis (PCA) 101, using R Peter Nistrup · Follow Published in Towards Data Science · 8 min read · Jan 29, 2019 2 Improving predictability and classification one dimension at a time! "Visualize" 30 dimensions using a 2D-plot! Basic 2D PCA-plot showing clustering of "Benign" and "Malignant" tumors across 30 features.
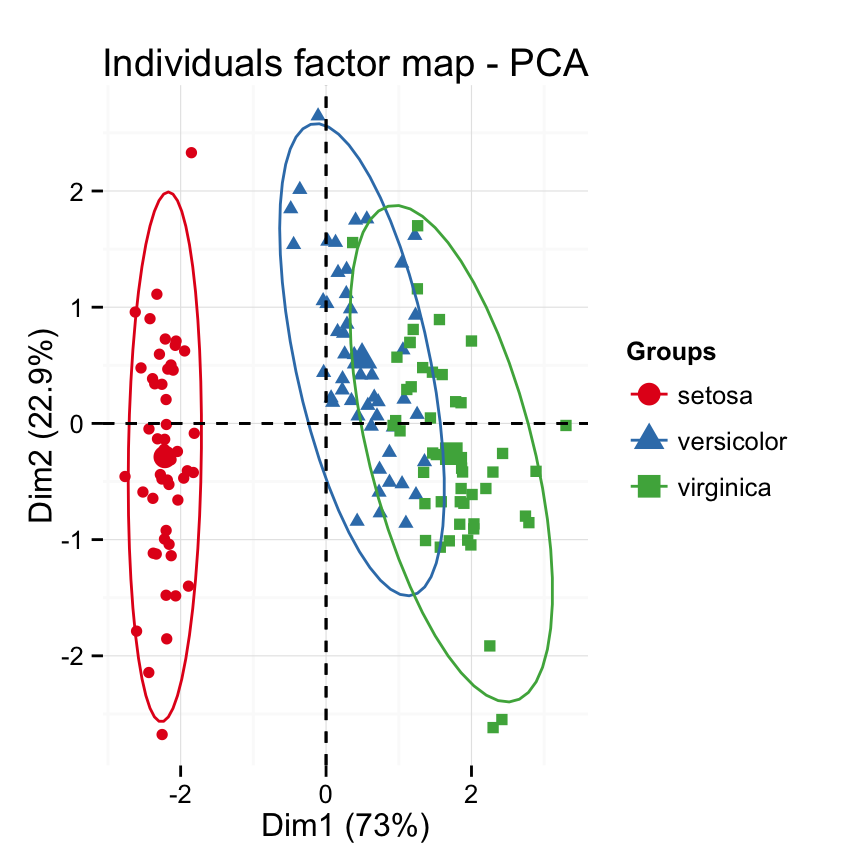
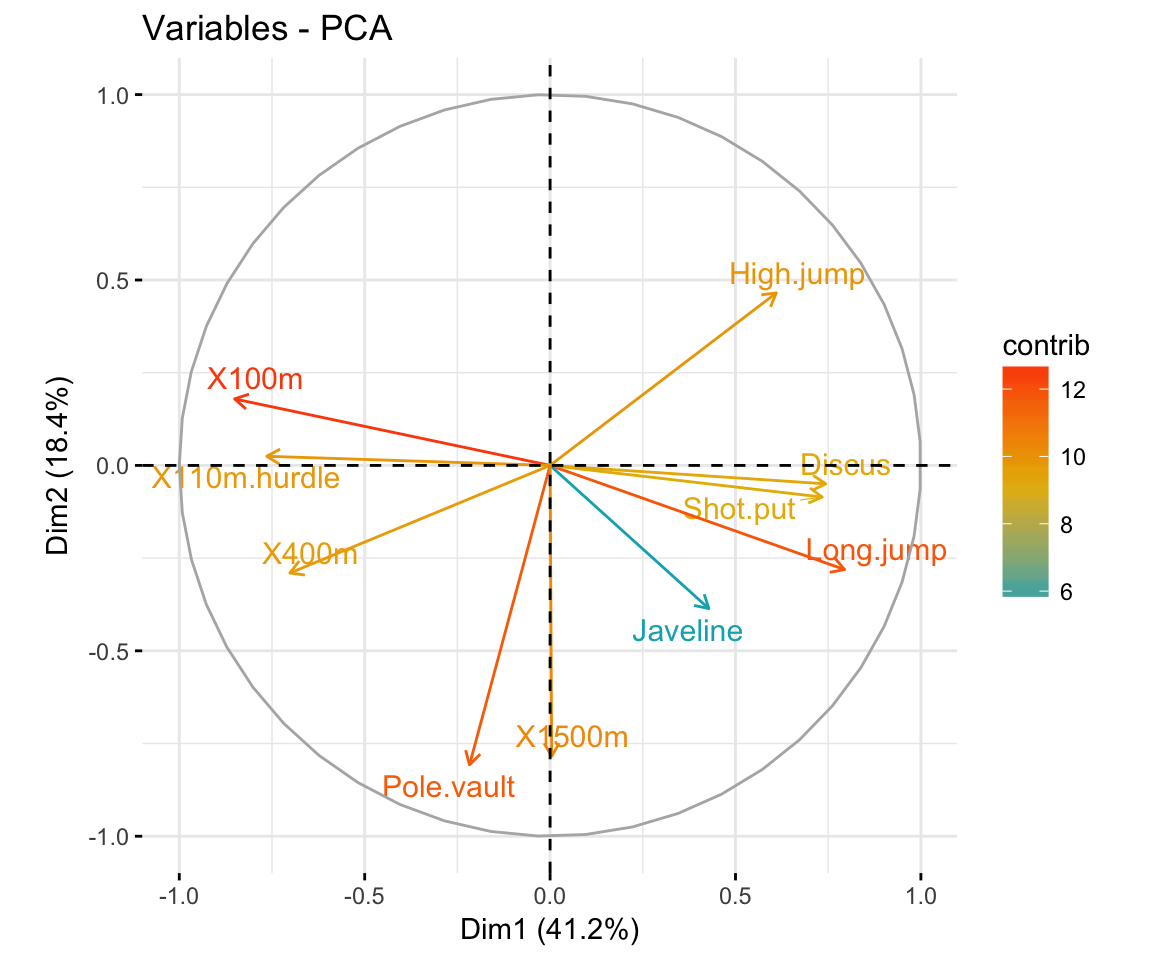
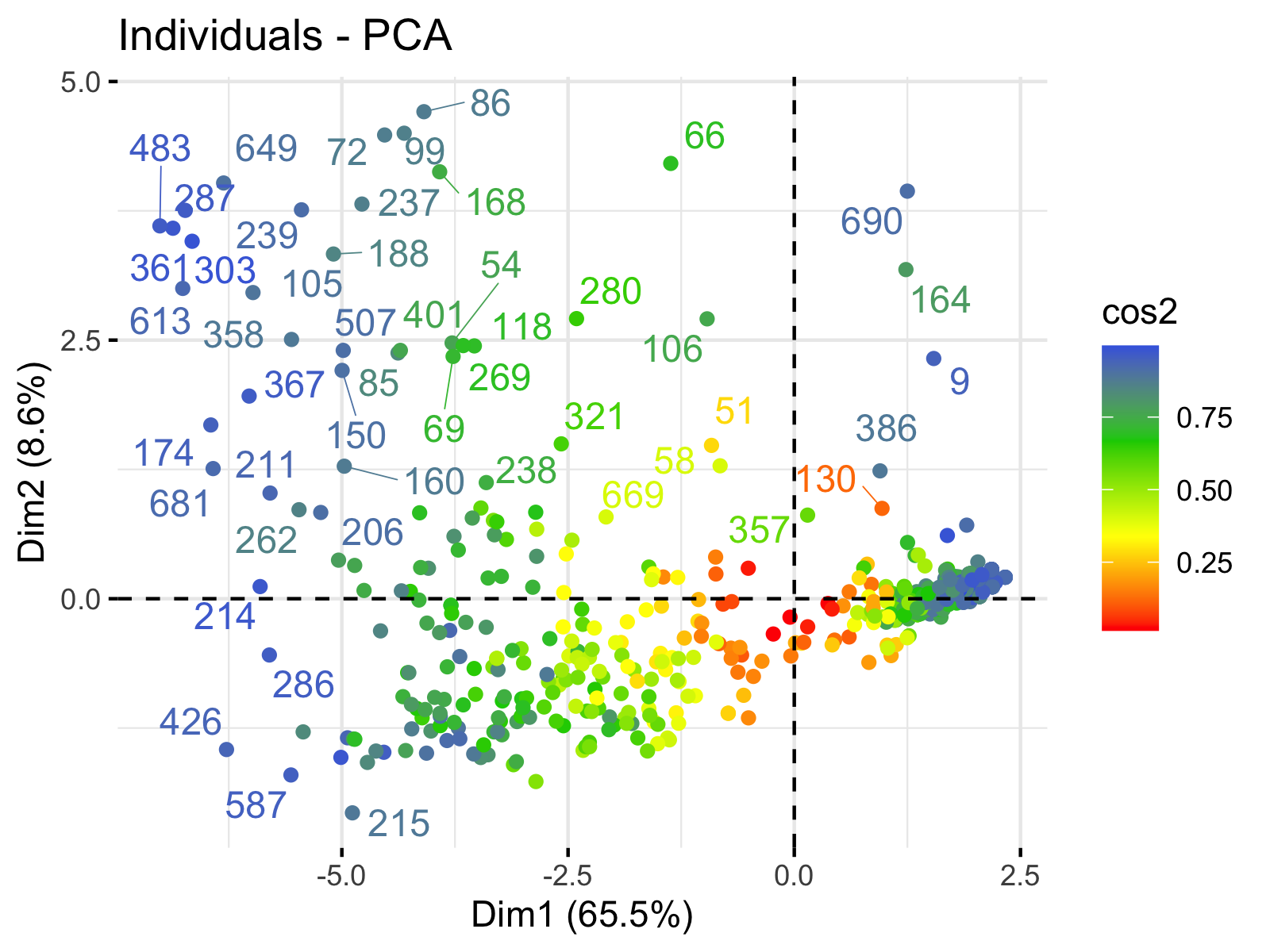
fviz_pca Quick Principal Component Analysis data visualization R software and data mining
Principal Component Analysis (PCA) is a widely-used statistical technique in the field of data science and machine learning. This article provides a step-by-step guide on implementing PCA in R, a popular programming language among statisticians and data analysts.

Principal Component Analysis (PCA) in R YouTube
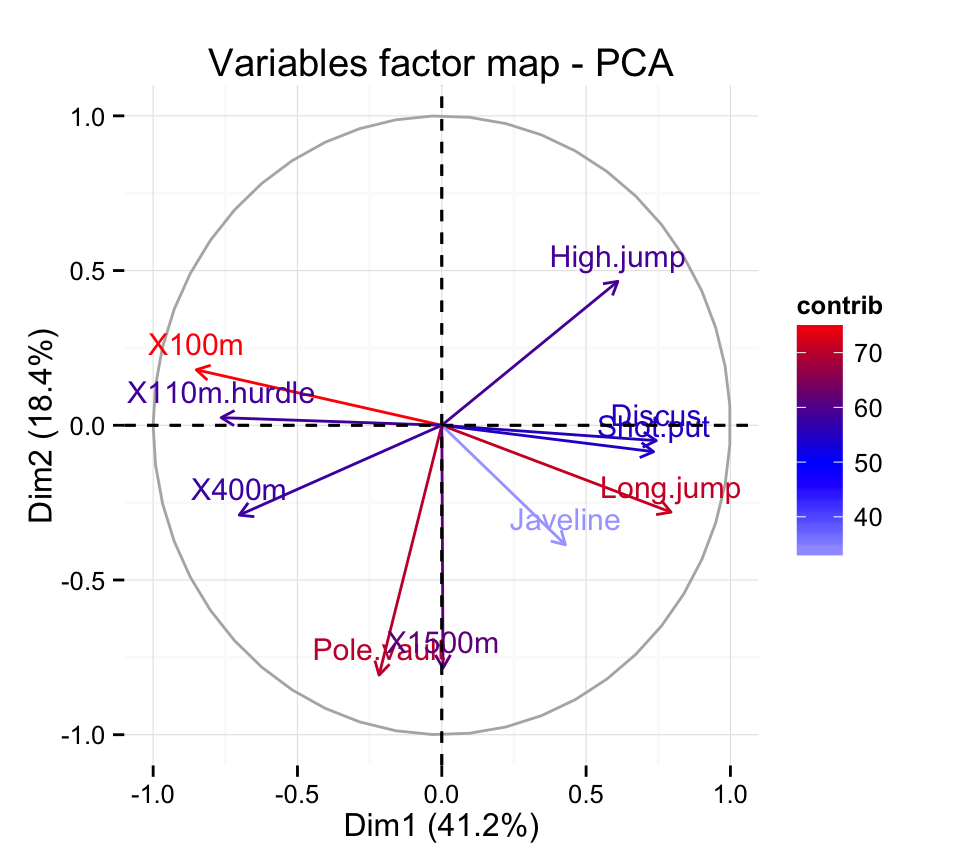
Principal component analysis ( PCA) allows us to summarize and to visualize the information in a data set containing individuals/observations described by multiple inter-correlated quantitative variables. Each variable could be considered as a different dimension.
GraphPad Prism 10 Statistics Guide Graphs for Principal Component Analysis
Feb 15, 2018. Principal Component Analysis (PCA) is unsupervised learning technique and it is used to reduce the dimension of the data with minimum loss of information. PCA is used in an application like face recognition and image compression. PCA transforms the feature from original space to a new feature space to increase the separation.
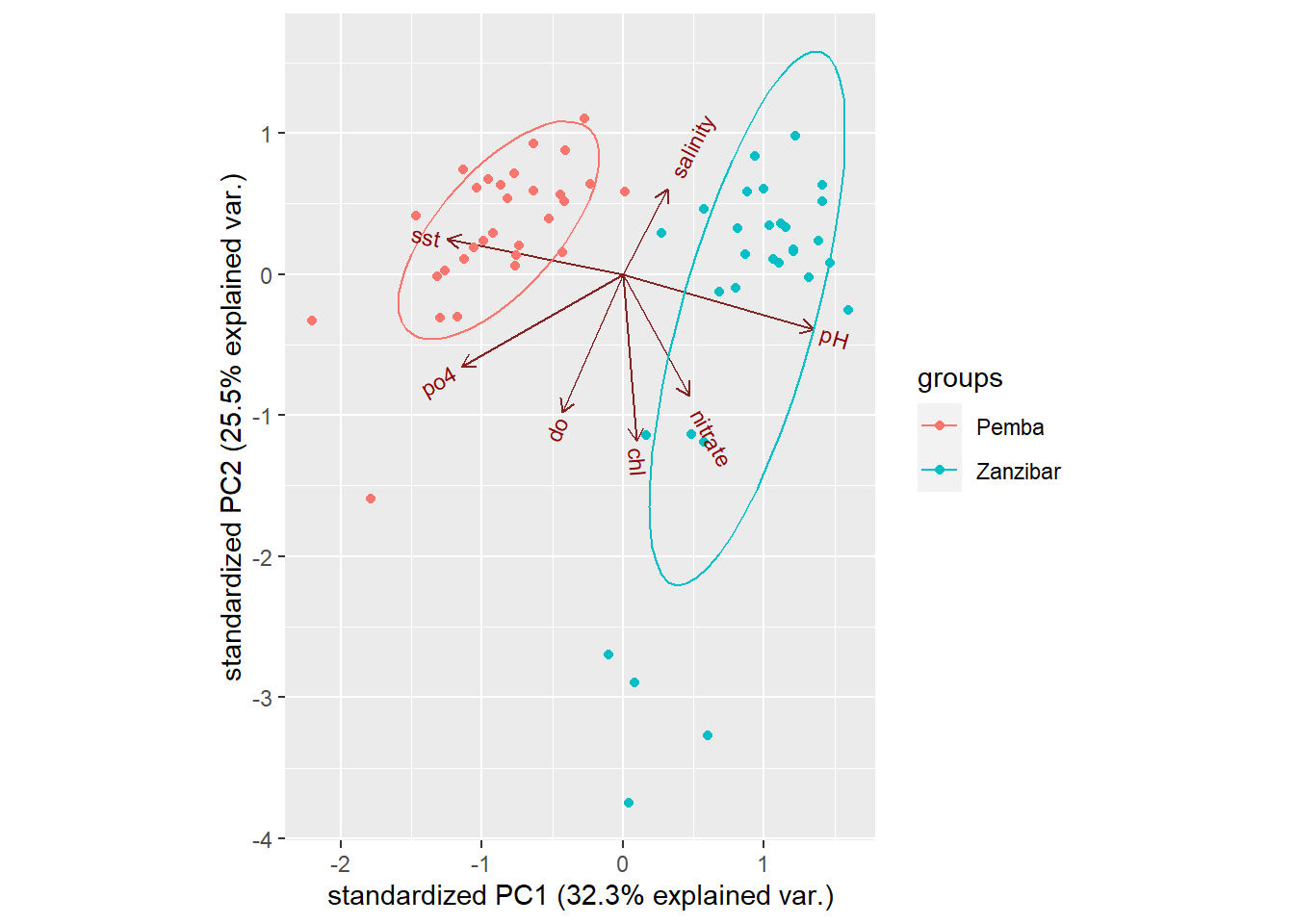
A simple Principal Component Analysis (PCA) in R Masumbuko Semba's Blog
In this tutorial, you will learn different ways to visualize your PCA (Principal Component Analysis) implemented in R. The tutorial follows this structure: 1) Load Data and Libraries 2) Perform PCA 3) Visualisation of Observations 4) Visualisation of Component-Variable Relation 5) Visualisation of Explained Variance